In silico Evaluation of Potential Bioactive Peptides of Phycoerythrins from Selected Rhodophytes
Abstract
Background
Rhodophytes typically possess a higher protein content than Chlorophytes and members of Class Phaeophyceae. The predominant proteins in rhodophytes are phycobiliproteins, constituting up to 50% of the overall protein composition. Phycoerythrin, phycocyanin, allophycocyanin, and phycoerythrocyanin are the principal phycobiliproteins. The objective of the study is to identify and test the activity of putative bioactive peptides derived from selected identical sequences of Rhodophytes. This study employs an in silico methodology to examine identical sequences of phycoerythrin from several rhodophytes as potential bioactive peptide precursors
Methods
in silico modeling of proteolysis was conducted utilizing papain, bromelain, thermolysin, pepsin, trypsin, and chymotrypsin A. Various bioinformatics tools, including PeptideRanker, PepCalc, ToxinPred, and AllerTop, were employed to assess the properties of the peptides
Results
The simulation revealed that the inhibitory effects of dipeptidyl peptidase IV (DPP IV) and angiotensin-converting enzyme (ACE) had the greatest potential. Peptides that inhibit alpha-glucosidase also exhibited certain efficacy. Stem bromelain had superior efficacy in hydrolysis percentages. This work illustrates those particular identical sequences, including phycoerythrin protein from rhodophytes may serve as a feasible natural alternative to synthetic ACE inhibitor medications. The research also indicates that the peptides may be advantageous in treating type 2 diabetes mellitus due to the existence of dipeptidyl peptidase IV peptides. Additionally, the analysis identified several novel peptides that may exhibit advantageous bioactivities
Conclusion
This study explored a prospective alternative supply of bioactive peptides in the food and pharmaceutical industry. These results establish a foundation for subsequent in vitro and in vivo investigations
1. INTRODUCTION
Rhodophytes, or red algae, represent a rich and intriguing assemblage of photosynthetic organisms that have engaged the interest of scientists and researchers for decades. These distinctive eukaryotic organisms are distinguished by their vibrant coloration, spanning deep reds to purples, and their capacity to flourish in many aquatic habitats [1, 2]. A distinguishing characteristic of rhodophytes is their varied pigmentation, mostly attributed to phycobiliproteins, including phycoerythrin and phycocyanin. These algae inhabit a variety of severe habitats, including shallow coastal waters, the deep ocean, and freshwater ecosystems [3]. Besides its ecological importance, rhodophytes have attracted interest for their possible commercial uses. These algae represent a feasible and cost-effective biomass supply of important chemicals, including phytopigments, polyunsaturated fatty acids, and numerous secondary metabolites [4]. They exhibit intriguing biological activity, positioning rhodophytes as a viable target for developing nutraceuticals, medicines, and other commercial products [2].
A light-harvesting pigment-protein complex known as phycoerythrin is present in red algae and certain cyanobacteria, providing a critical function in photosynthesis. The algae obtained their red coloration from phycoerythrin, with R-phycoerythrin being the predominant protein present (Fleurence, 2003; Glazer, 1984). This protein consists of two primary subunits, the α and β subunits, which collectively constitute the functional structure [5] and indicates a molecular weight between 240 and 260 kDa [6]. There are also some exceptional instances. One of these proteins consists of alpha and beta chains with a gamma linker ((αβ)6γ), as detected in electron density maps from Gracilaria chilensis [7]. Phycoerythrins are comparatively stable and water-soluble proteins [8]. Besides its role in light absorption, phycoerythrin has been studied for potential bioactive effects. R-Phycoerythrin from Spyridia filamentosa and Fusco-purpura exhibits significant antioxidant activity [9, 10] and inhibits angiotensin I-converting enzyme through the production of bioactive peptides [11]. It has demonstrated antioxidant, anti-inflammatory, and possibly anticancer properties, rendering it a significant focus for further investigation and prospective uses in the nutraceutical, cosmetic, and pharmaceutical sectors [12]. The distinctive characteristics of phycoerythrin, along with the rising demand for sustainable and natural sources of colorants and bioactive chemicals, have become an area of increasing investigation in science [1, 12, 13].
Bioactive peptides remain dormant within the sequence of the parent protein yet possess the capability to enhance human health [14, 15]. The selection of enzyme and hydrolysis conditions can profoundly influence the resultant peptide profile and its biological characteristics [16]. Bioactive peptides obtained from rhodophytes have attracted considerable interest due to their various therapeutic qualities, including antioxidant, anti-inflammatory, antibacterial, and anticancer activity [17]. The enzymatic hydrolysis of phycoerythrin has garnered attention due to its potential for generating beneficial peptides and amino acids with biological activity.
Proteomics advances have led to the creation of in silico proteolysis tools like BIOPEP (www.uwm.edu.pl/biochemia/index.php/pl/biopep), which can predict the production of bioactive peptides using a combination of proteases with particular cleavage sites and varied substrates using known protein sequences. Additionally, in silico proteomic techniques can characterize vast numbers of proteins or protein fragments from complex dietary items and create profiles of probable biological activity [18, 19]. In this research, BIOPEP was used to determine probable protein fragments, quantify the value of proteins (in terms of potential biological activities and frequency of release of bioactive peptides), and predict which links in a protein chain are likely to be hydrolyzed by endopeptidases. The objective of this in silico proteolysis technique is to identify the bioactive peptides of phycoerythrin alpha and beta subunit from red algae for their potential pharmaceutical use.
2. MATERIALS AND METHODS
The National Centre for Biotechnology Information (NCBI) database contains 5,394 phycoerythrin protein sequences, including 408 identical proteins derived from red algae (accessed on 20th March 2024). The Identical Protein Groups repository comprises a singular entry for each protein translation identified across multiple NCBI sources, including annotated coding sections in GenBank and RefSeq, along with records from SwissProt and PDB. This resource enables researchers to achieve more precise search outcomes and swiftly find a specific protein of interest. In the investigation, the identical sequences were chosen based on the maximum quantity of lie sequences. The study identifies the identical sequences of access number YP_008965769.1 (R-phycoerythrin alpha subunit) protein, which comprises 1552 phycoerythrin protein sequences from Rhodophyta. Correspondingly, the identical proteins with accession numbers NP_053978.1 (phycoerythrin alpha subunit), YP_009313668.1 (phycoerythrin subunit b), and YP_009294675.1 (phycoerythrin beta subunit) comprise 45, 12, and 9 sequences, respectively, from various sources of red algae. Multiple sequence alignments were applied using ClustalOmega (available at https://www.ebi.ac.uk/Tools/msa/clustalo/). The sequence homology was identified by UniProtKB/Swiss-Prot database.
The BIOPEP-UWM database was utilized to predict bioactive peptides, with the 'Profiles of potential biological activity' action menu calculating the number of probable peptides. The frequency of bioactive fragment occurrence of phycoerythrin protein sequence was calculated using the following equation eq. (1).
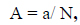
where a is the number of fragments with given activity in a protein sequence, and N is the number of amino acid residues of the protein. The parameter 'A' in protein assessment measures the bioactivities of peptides encoded in a protein sequence, allowing for quick identification of bioactive fragments. However, it does not reveal specific sequence motifs or their location in the protein chain, which is the qualitative characteristic for protein evaluation known as the profile of potential biological activity of a protein [20]. The total frequency of bioactive fragments of each phycoerythrin protein sequence (∑A) was calculated, encompassing the frequency of occurrence for all activities provided by the selected proteins. In this study, A1 and A2 denote the bioactivities of ACE and DPP IV inhibitors, respectively. A3 represents the sum of all other bioactivities.
The potential biological activity refers to the protein's beneficial effects (such as ACE inhibitor, alpha-glucosidase inhibitor, DPP IV inhibitor, etc.). The potential biological activity of each protein, B (mM–1), was calculated using the equation eq. (2).
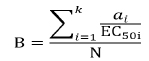
Where ai is the number of times the i-th bioactive fragment appears in the protein sequence, EC50i is the concentration of the i-th bioactive peptide that corresponds to its half-maximal activity (µM) or half-maximal inhibition (IC50) in the case of peptides with inhibitory activity, k is the number of different fragments with a given activity, and N is the number of amino acid residues. “B” is a key factor in determining a protein's biological activity, referring to the number of fragments with specific biological activity and its half-maximal inhibition. A low number of active fragments or weak peptides can result in zero or inconsequential values. A high number of fragments indicates a specific biological activity, while zero indicates some fragments may be active but not enough.
The enzyme action tool of BIOPEP-UWM was chosen to do artificial proteolysis because it can represent bioactive peptides after cleavage with each enzyme (http://www.uwm.edu.pl/biochemia). The bioinformatics tool was developed by Minkiewicz et al. [19]. Papain, thermolysin, stem bromelain, pepsin, trypsin, and chymotrypsin A were independently used against the substrate protein sequences to release peptides. Thermolysin cleaves bulky and aromatic residues (Ile, Leu, Val, Ala, Met, Phe) in position P1′ [21, 22]. and the peptides produced by thermolysin have a lot of promise as ACE inhibitor peptides [22]. Papain has been discovered to be helpful in the production of ACE inhibitors and antioxidative peptides in the literature [23-26]. Following chymotrypsin C, papain created the most bioactive peptides in silico analysis of patatin with 16 enzymes [26]. These food-grade enzymes are also available commercially. These food-processing proteases were chosen in conjunction with gastrointestinal enzymes for these reasons.
The frequency of release of peptides with given bioactivity by selected enzymes, or “frequency of release” (AE), and the relative frequency of release of peptides with given activity by selected enzymes (W) were computed using the following equations to estimate the efficiency of bioactive fragment release eq. (3,4)
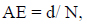
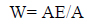
Where N is the number of amino acid residues in the protein and d is the number of peptides with a given activity produced from the protein sequence by the selected enzyme.
BIOPEP-UWM returns the fragments associated with the database action. The activities and EC50 value of the known peptides were identified from the tool and related literature. Inhibitors of dipeptidyl peptidase IV (DPP-IV) are typically reported to contain short amino acid sequences of less than five amino acids [27]. Hall et al. (2018) proved that small molecular weighted peptides had the highest DPP IV inhibitory activity [28]. Similarly, tropical blended cricket powder showed improved DPP-IV inhibitory activity after simulated gastrointestinal digestion [29]. Therefore, most of the DPP-IV inhibitory peptides were identified as di or tripeptides [30, 31]. Other dietary proteins, such as salmon skin gelatin [32] and quinoa protein hydrolysate [26], showed the same pattern, with lower molecular weight peptides having stronger DPP IV inhibitory action. As a result, to look for novel peptides after enzymolysis with the chosen enzyme, the fragments were ranked for potential activity as previously described [27]. The bioinformatics tool Peptidranker was used to predict the potential novel peptide. This tool can assess the likelihood of a peptide being bioactive based on structural analysis [33].
In addition, in silico analysis was used to estimate probable peptide properties. Water solubility, digestion resistance, toxicity, and allergenicity were all tested. PepCalc, which can be found at http://pepcalc.com, was used to predict solubility in water. PeptideCutter, accessible at http://web.expasy.org/peptide_cutter, was used to predict gastrointestinal digestion. Chymotrypsin-low specificity, chymotrypsin-high specificity, pepsin (pH 1.3), pepsin (pH > 2), and trypsin enzymes were employed to measure resistance [27]. ToxinPred and AllerTOP, available at http://www.imtech.res.in/raghava/toxinpred/ and http://www.pharmfac.net/allertop, were used to assess toxicity and allergenicity.
3. RESULTS
3.1. Sequence Analysis
By using Clustal Omega, multiple sequence alignment of amino acid sequences of phycoerythrin alpha and beta subunit from rhodophyte. Two different identical protein sequences each of alpha (Porphyridium purpureum vs Porphyra purpurea) and beta (Helminthocladia australis vs Gracilariopsis chorda) subunit of phycoerythrins revealed (Fig. 1-2) variable pairwise sequence similarities or homology with the percent identity matrix 87.20 and 92.66. A study to examine the RuBisCO (LS) among different Caulerpa species found alignment sequence difference was 92-100% [34].

Sequence alignment of porphyridium purpureum vs porphyra purpurea

Sequence alignment of helminthocladia australis vs gracilariopsis chorda.
3.2. Generated profile and prediction of bioactive peptides
Within the in silico hydrolysis products released from phycoerythrin proteins of rhodophytes bioactive peptides were identified using the BIOPEP_UWM. On 20th March 2024, 4800 peptides formed in 74 bioactivities were found in the BIOPEP-UWM database. From selected proteins, fragments were released with biological activities such as: ace inhibitor, activating ubiquitin-mediated proteolysis, alpha-glucosidase inhibitor, antiamnestic, antibacterial, anticancer, antioxidative, antithrombotic, anti-inflammatory, antiviral, bacterial permease ligand, campde inhibitor, dipeptidyl peptidase iii inhibitor, dipeptidyl peptidase iv (dpp iv) inhibitor, haemolytic, hypotensive, hypolipidemic, leucyltransferase inhibitor, lactocepin inhibitor, neuropeptide, protein associated with Myc (pam) inhibitor, regulating, renin inhibitor, stimulating, toxic and xaa-pro inhibitor. Peptides were labeled as “regulatory” that were involved in the activation of stomach mucosa membrane and/or phosphatase and kinase, as well as the regulation of cell permeability, ion flow, heart muscle contraction, and mechanism of phosphoinositol action. Peptides with one of the aforementioned activities were infrequently found. Typically, these activities possess a parameter A value of less than 0.300 [20]. Stimulating fragments in the study relate to the peptides that stimulate the release of vasoactive substances and glucose uptake [35, 36]. Table 1 shows the prominent number of active fragments as well as their potential biological activities, B (mm–1), where appendix (Table A1) shows the detail: A value from the sequences of Porphyridium purpureum, Porphyra purpurea, Helminthocladia australis and Gracilariopsis chorda. The analysis showed the phycoerythrins released the highest numbers of dpp iv inhibitor peptide and ace inhibitor fragments with potential biological activity. There were some alpha-glucosidase inhibitor activities for three identical sequences, but the renin inhibitor showed activity only for gracilariopsis chorda identical sequences. The highest B value was found to be 0.0213- 0.0064 (ACE inhibitor) and the lowest was 0 or negligible. Other active fragments did not show any results when calculating their b values. The result is constant with in silico evaluation of seagrass halophila stipulacea. The bioactive antioxidative fragments of rubisco protein showed B values 0 by enzymatic hydrolysis [37].
3.2.1. Frequency of Occurrence of Peptides from Phycoerythrins from Study Sequences
The total frequency of occurrence of all existing activities (∑A) of the Phycoerythrins varies from 1.5124 to 1.316. Identical phycoerythrin alpha subunit sequences of Porphyridium purpureum gave the highest value and phycoerythrin beta subunit sequences of Gracilariopsis chorda the lowest. ACE inhibitor, activating ubiquitin-mediated proteolysis, alpha-glucosidase inhibitor, antiamnestic, antioxidative, antithrombotic, dipeptidyl peptidase III inhibitor, dipeptidyl peptidase IV inhibitor, hypotensive, neuropeptide, regulating, renin inhibitor, stimulating, PAM inhibitor were the widespread activities of all studied sequences. Therefore, the identical sequences containing all the species possess the activities. These data demonstrated an apparent distinction of these species to use in nutraceutical product development. Table 2 also quantified that ACE and DPP IV inhibitors were the major parts of the occurrence of bioactive fragments and according to biological activities (B), ACE, DPP IV and Alpha-glucosidase inhibitors showed potential activity. Thus, phycoerythrins are predicted to be potential for the release of these peptides. It showed all the identical sequences from different species are mostly potential for ACE and DPP IV inhibition.
3.2.2. Frequency of Release of Fragments with a given Activity by Selected Hydrolysis Enzymes
Table 3 shows the theoretical degree of hydrolysis and released number of potential fragments by the action of different enzymes. Stem bromelain showed the highest degree of hydrolysis and papain, thermolysin can release the highest number of potential peptides. It is shown that all the identical sequences from different species appear almost the same despite being divergence.
In stem bromelain, the hydrolysis percentage was higher, and it showed the highest efficiency for releasing ace and dpp iv inhibitory peptides and it is factual for all the identical protein sequences (Table 4).
3.3. In-silico proteolytic hydrolysis of sequences of phycoerythrins
Table 5 shows the phycoerythrins that can release bioactive peptides that have proven beneficial effects. Most of the bioactive peptides are Angiotensin I converting enzyme inhibitors and/or Dipeptidyl peptidase IV inhibitors as expected. These peptides listed with their specific bioactivities demonstrated their significance in therapeutics.
Amino acids are represented using single-letter codes. A-Alanine,C- Cystine, D-Aspartic acid, E-Glutamic acid, F-Phenylalanine, G-Glycine, I-Isoleucine, K- Lysine, L-Leucine, M-Methionine, N-Asparagine, P-Proline, Q-Glutamine, R-Arginine, S-Serine, T-Threonine, V-Valine, Y-Tyrosine. ah-Angiotensin I converting enzyme inhibitor, glui-Alpha-glucosidase inhibitor, dpp-Dipeptidyl peptidase IV inhibitor, dpp3-Dipeptidyl peptidase iii inhibitor, re- Regulating, ne-Neuropeptide, xaap-XAA-pro inhibitor, hypl-Hypolipidemic, lcp-Lactocepin inhibitor ren-Renin inhibitor, ao- Antioxidative, 35pd- CaMPDE inhibitor.
| Activity | Identical Protein Sequence Organism (NCBI accession number) | |||
|---|---|---|---|---|
|
P. purpureum (YP_ 008965769.1) |
P. purpurea (NP_ 053978.1) |
H. australis (YP_ 009313668.1) |
G. chorda (YP_ 009294675.1) |
|
| Number of active fragments (Potential biological activity, B) | ||||
| ah | 82 (0.017375) |
84 (0.0213637) | 79 (0.009839736) | 73 (0.006434771) |
| glui | 6 (1.95E-06) | 6 (1.95E-06) | 3 (2.20E-07) | 2 |
| dpp | 102 (0.000164841) |
103 (0.000168671) | 111 (0.000295123) | 102 (0.000220363) |
| ren | 2 | 2 | 6 | 5 (1.82E-06) |
| Species |
Activities (Total number) |
∑A | A1 | A2 | A3 |
|---|---|---|---|---|---|
| P. purpureum | Widespread activities, ai, 35pd, hypl, lcp, xaap (19) | 1.5124 | 0.5183 | 0.622 | 0.3721 |
| P. purpurea | Widespread activities, 35pd, ai (16) | 1.4814 | 0.5122 | 0.628 | 0.3416 |
| H. australis | Widespread activities, ab, ac, 35pd, leut, tox, lig, he, avi (22) | 1.4571 | 0.4463 | 0.6271 | 0.3837 |
| G. chorda | Widespread activities, ab, ac, avi, he, leut (19) | 1.316 | 0.4124 | 0.5763 | 0.3273 |
| Enzyme | Identical Protein Sequence Organism (NCBI accession number) | |||
|---|---|---|---|---|
|
P.purpureum DHt (Fr) |
P. purpurea DHt (Fr) |
H. australis DHt (Fr) |
G. chorda DHt (Fr) |
|
| Papain | 41.81 (44) | 39.87 (40) | 41.48 (47) | 38.64 (47) |
| Stem Bromelain | 55.82 (42) | 57.67 (41) | 57.66 (40) | 57.67 (40) |
| Thermolysin | 42.94 (42) | 41.72 (44) | 41.72 (44) | 41.72 (44) |
| Pepsin (PH 1.3) | 8.82 (16) | 9.20 (15) | 9.20 (15) | 9.20 (15) |
| Trypsin | 10.43 (18) | 10.42 (18) | 10.42 (17) | 10.42 (18) |
| Chymotrypsin A | 25.15 (31) | 23.31 (29) | 23.31 (29) | 23.31 (29) |
In silico proteolysis of phycoerythrin sequences released various peptides with multiple activities. Table 6 shows the common peptides with multiple activities from all the selected phycoerythrin sequences. These peptides demonstrate their unique possibility for drug development.
3.3.1. Predicted Potential Characteristics of Novel Peptides
Table 7 shows the parameters of the predicted potential novel peptides from all selected identical protein sequences. These data indicated that all the peptides identified in this table may be bioactive according to their probability score. These characteristics can inform future investigations on these peptides. The novel peptide AC possesses a unique characteristic of resistance to digestion. It was also anticipated that the other peptides would be degraded by gastrointestinal digestive enzymes; however, this could be circumvented through techniques such as encapsulation [55]. All the peptides were probably nontoxic, according to the ToxinPred analysis.
| Enzyme | Identical Protein Sequence Organism | |||||
|---|---|---|---|---|---|---|
| P. purpureum | P. purpurea | H. australis | G. chorda | |||
| ACE inhibitory peptides | Papain | AE | 0.072 | 0.079 | 0.039 | 0.039 |
| W | 0.141 | 0.159 | 0.089 | 0.089 | ||
| Stem Bromelain | AE | 0.104 | 0.085 | 0.085 | 0.085 | |
| W | 0.200 | 0.167 | 0.167 | 0.167 | ||
| Thermolysin | AE | 0.037 | 0.031 | 0.031 | 0.031 | |
| W | 0.070 | 0.059 | 0.059 | 0.059 | ||
| Pepsin (PH 1.3) | AE | - | 0.006 | 0.006 | 0.006 | |
| W | - | 0.012 | 0.012 | 0.012 | ||
| Trypsin | AE | 0.006 | 0.006 | 0.006 | 0.006 | |
| W | 0.012 | 0.012 | 0.012 | 0.012 | ||
| Chymotrypsin A | AE | 0.018 | 0.024 | 0.024 | 0.024 | |
| W | 0.035 | 0.047 | 0.047 | 0.047 | ||
| DPP IV inhibitory peptides | Papain | AE | 0.084 | 0.079 | 0.090 | 0.079 |
| W | 0.137 | 0.126 | 0.144 | 0.137 | ||
| Stem Bromelain | AE | 0.091 | 0.085 | 0.085 | 0.085 | |
| W | 0.147 | 0.136 | 0.136 | 0.136 | ||
| Thermolysin | AE | 0.055 | 0.048 | 0.048 | 0.048 | |
| W | 0.088 | 0.078 | 0.078 | 0.078 | ||
| Pepsin (PH 1.3) | AE | 0.006 | 0.012 | 0.012 | 0.012 | |
| W | 0.009 | 0.019 | 0.019 | 0.019 | ||
| Trypsin | AE | 0.012 | 0.012 | 0.012 | 0.012 | |
| W | 0.019 | 0.019 | 0.019 | 0.019 | ||
| Chymotrypsin A | AE | 0.031 | 0.031 | 0.031 | 0.031 | |
| W | 0.049 | 0.049 | 0.049 | 0.049 | ||
| Porphyridium Purpureum | ||||
|---|---|---|---|---|
|
Activities Enzyme |
ah | dpp | glui | others |
| Papain | AG, AR, DR, PL, PT, QG | AD, AG, AL, DR, IN, NL, PL, PT, QG | AD | PL (xaap, lcp) |
| Bromelain | CF, DA, DL, DR, EA, EF, EV, IA, PL, PR, PT, YL, YV | ES, EV, IA, NL, PL, PS, PT, QA, YL, YR, YT, YV | EA | PL (xaap, lcp), DA, PR, YL, YR (dpp3), EF (hypl, ren, 35pd), YL, YR (ne) |
| Thermolysin | AG, AR, ITT, LN, VE | AD, AG, AS, IN, LN, VE, VN, VS | AD, VE | - |
| Pepsin | - | NL | - | - |
| Trypsin | DR | DR, MK | - | - |
| Chymotrypsin | AY, DY, RL | AL, AY, IN, RL, VN | AY (ao), DY (re) | |
| Porphyra purpurea | ||||
| Papain | AF, AG, AR, DR, PT, QG | AD, AF, AG, DR, NL, PT, QG | AD | - |
| Bromelain | CF, DA, DL, DR, EA, EV, IA, PR, PT, QG, YL, YV | DR, ES, EV, IA, NL, PS, PT, PV, QA, QG, YL, YR, YV | EA | DA, PR, YL, YR (dpp3), YL, YR (ne) |
| Thermolysin | AG, LN, VE | AD, AG, AS, LN, VE, VN, YS | AD, VE | - |
| Pepsin | AF | AF, NL | - | - |
| Trypsin | DR | DR, MK | ||
| Chymotrypsin | AF, DY, RL, SY | AF, RL, SL, SY, VN | DY, SL (re) | |
| Helminthocladia australis | ||||
| Papain | AF, AG, AV, DG, NG, SG | AF, AG, AL, APG, AS, AV, ML, NG, NR, NT, YI | - | NR (ren), YI (dpp3) |
| Bromelain | CF, DA, DL, DR, EA, EV, IA, PR, PT, QG, YL, YV |
DR, ES, EV, IA, NL, PS, PT, PV, QA, QG, YL, YR, YV | EA | DA, PR, YL, YR (dpp3), YL, YR (ne) |
| Thermolysin | AG, AR, LN, VE | AD, AG, AS, LN, VE, VN, YS | AD, VE | - |
| Pepsin | AF | AF, NL | - | - |
| Trypsin | DR | DR, MK | - | - |
| Chymotrypsin | AF, DY, RL, SY | AF, RL, SL, SY, VN | DY, SL (re) | |
| Gracilariopsis chorda | ||||
| Papain | AF, AG, AV, DG, NG, SG |
AF, AG, AL, AS, AV, KT, ML, NG, NR, YI | - | NR (ren), YI (dpp3) |
| Bromelain | CF, DA, DL, DR, EA, EV, IA, PR, PT, QG, YL, YV |
DR, ES, EV, IA, NL, PS, PT, PV, QA, QG, YL, YR, YV | EA | DA, PR, YL, YR (dpp3), YL, YR (ne) |
| Thermolysin | AG, AR, LN, VE | AD, AG, AS, LN, VE VN, YS |
AD, VE | - |
| Pepsin | AF | AF, NL | - | - |
| Trypsin | DR | DR, MK | - | - |
| Chymotrypsin | AF, DY, RL, SY | AF, RL, SL, SY, VN | - | DY, SL (re) |
| Peptide/ Enzyme/Refs. | EC50 (µM) | |||
|---|---|---|---|---|
| Papain | Activity | ACE inhibitory | DPP inhibitory | Other activities |
| AG [38, 39] DR [39, 40] |
ah, dpp | 2500 110.50 |
0.00 0.00 |
- |
| Stem bromelain | ||||
| DA [41, 42] | ah, dpp3 | 3800 | - | 0.00 (dpp3) |
| DR [39, 40] EV [39, 43] IA [44, 45] PT [39, 43] YV [39, 46] |
ah, dpp | 110.50 0.00 153.00 0.00 575.50 |
0.00 0.00 0.00 0.00 0.00 |
|
| YL [39, 47-50] | ah, dpp, dpp3, ne, ao | 0.00 | 0.00 | 0.00 (dpp3), 0.00 (ne), 0.00 (ao) |
| EA [38, 51] | ah, glui | 100000 | 17000 | |
| Thermolysin | ||||
| AD [39, 51] | glui, dpp | - | 0.00 | 25660 (glui) |
| AG [38, 39] LN [39, 43] |
ah, dpp | 2500 0.00 |
0.00 0.00 |
|
| VE [39, 43, 51] | ah, dpp, glui | 0.00 | 0.00 | 22170.00 |
| Trypsin | ||||
| DR [39, 40] | ah, dpp | 110.50 | 0.00 | |
| Chymotrypsin | ||||
| DY [46, 52] | ah, re | 100 | 0.00 (re) | |
| RL [53, 54] | ah, dpp | 2439.00 | 0.00 | |
| Peptides | Probability Score | Allergenicity Probability | Resistance to Digestion | Solubility in Water | Molecular Weight (g/mole) |
|---|---|---|---|---|---|
| Stem Bromelain | |||||
| NYCL | 0.736 | Allergen | No | Poor | 511.59 |
| DHYMR | 0.637 | Allergen | No | Good | 720.80 |
| DEWG | 0.516 | Not Allergen | No | Good | 505.48 |
| Thermolysin | |||||
| LC | 0.840 | Allergen | No | Poor | 234.32 |
| YMR | 0.785 | Not Allergen | No | Good | 468.57 |
| AC | 0.735 | Not Allergen | Yes | Poor | 192.24 |
| AGR | 0.548 | Not Allergen | No | Good | 302.33 |
| Pepsin | |||||
| VNYCL | 0.540 | Allergen | No | Poor | 610.72 |
| Trypsin | |||||
| CRY | 0.793 | Not Allergen | No | Good | 440.52 |
| EAGDACFAK | 0.553 | Not Allergen | No | Good | 910.99 |
| LCVPR | 0.511 | Allergen | No | Good | 586.75 |
| Chymotrypsin | |||||
| CL | 0.879 | Allergen | No | Poor | 234.32 |
| CVPRDM | 0.601 | Not Allergen | No | Good | 719.88 |
| KCY | 0.520 | Allergen | No | Good | 412.51 |
4. DISCUSSION
Marine algae have been found to contain a diverse array of primary and secondary metabolites that exhibit a wide range of biological activities, making them a promising target for the discovery of novel bioactive compounds [56]. Leveraging the vast protein sequences found in algal genomes has become an area of increasing research focus as scientists seek to uncover potentially bioactive peptide fragments that could possess valuable therapeutic properties [56-59].
Enzymatic hydrolysis is the most common mechanism for releasing biologically active peptides [60]. A recent study showed that enzymatic hydrolysis can improve the extraction of R-phycoerythrin, proteins, and sugar (water-soluble component) from Gracilaria gracilis [61]. The degree of hydrolysis by enzymes is crucial in the formation of bioactive peptides. In vitro studies show that the secondary structure of a protein can affect its enzymic activity. For example, di-sulfide links in the structure of proteins make them more resistant to proteolysis [62]. However, because proteolysis is partly governed by cleavage sites available in the substrates, the degree of hydrolysis is not always proportional to protease concentration [63]. Antioxidant, antidiabetic, antihypertensive, antithrombotic, immuno-modulating, osteoprotective, antibacterial, anticarcinogenic, and growth-promoting effects have been revealed in peptides produced from dietary proteins by enzymatic hydrolysis [28]. From wheat gluten, bovine muscle proteins, patatin (potato tuber protein), and quinoa, papain has been shown to successfully synthesize ACE inhibitors and antioxidative peptides [34]. In a previous in silico analysis of Quinoa and soybean proteins, stem bromelain was found to have the greatest degree of hydrolysis [64]. An in vitro study showed that ACE-1 inhibitor and renin inhibitory peptides can be released by hydrolysis of papain in green algae Ulva lactuca [65]. in silico study on RiBisCO protein of Caulerpa spp. showed that a high number of bioactive peptides were obtained by papain proteolysis [34]. These studies indicated that papain and bromelain were more effective at releasing bioactive peptides from the breakdown of plant protein. According to this study, pepsin, trypsin, and chymotrypsin A were comparably less active and had a lower capacity to produce bioactive peptides than papain, bromelain, and thermolysin from selected rhodophytes (Table 3). Similarly, the effect of the enzymatic action of chymotrypsin, trypsin, and pepsin showed lower activity in sorghum, wheat, and rice. Besides, thermolysin and papain were highest in sorghum, and bromelain in wheat and rice showed the highest A score [66].
Bioinformatics is defined as “the application of computing resources to biological data” [67]. Bioinformatics has a long history in fields such as molecular medicine, comparative genomics, molecular evolution, microbial genome applications, and drug development. However, bioinformatics-based in silico methods have recently been applied to investigate bioactive peptides in proteins [67]. in silico investigations are also useful for identifying peptides with specific bioactivities, understanding their mechanisms of action, and learning more about their structural needs for bioactivity [68]. When it comes to the theoretical calculation of prospective bioactivities and the corresponding activity of protein after hydrolysis with specific proteases, the use of in silico technologies will save money and time. in silico methods are used to mine bioactive peptides at first, allowing researchers to concentrate on a limited number of peptide candidates that are most likely to have high potency of the desired biological activities [69]. For instance, Phe-Cys derived from in silico enzymolysis (thermolysin) of cereal crop RuBisCO (ls) was discovered to be potentially active because of its high value assigned by the PeptideRanker tool. A subsequent in vitro investigation revealed that this dipeptide possesses strong antioxidant properties [70, 71]. Lafarga et al. identified novel ACE and DPP-IV inhibitor peptides in bovine serum albumin, utilizing in silico technologies, including BIOPEP and PeptideRanker, in their research [72]. These data unequivocally demonstrate that in silico research can complement wet-lab in vitro enzyme action studies.
The bioactivity was predicted using the PeptideRanker in this study (Table 7). It is a bioinformatic tool that utilizes an N-to-1 neural network algorithm to predict and rank the probability of a peptide being bioactive. PeptideRanker assigns scores from 0 to 1. The higher the score, the more likely the peptide is to be bioactive. PeptideRanker was trained with a 0.5 threshold, meaning that any peptide predicted to be higher than this threshold is classified as bioactive. One of the tool's limitations is that it does not specify the most appropriate activities [33, 73, 74]. Similar to this study, novel anti-inflammatory and anti-cancer peptides were found through in silico analysis using PeptideRanked as a predictor [75]. Furthermore, PeptideRanker’s findings indicated that some di-, tri-, and/or polypeptides derived from the RuBisCO protein of the seaweed Ulva Lactuca possess significant promise as bioactive peptides [76].
The phycoerythrin from selected rhodophytes in the present study exhibits a greater quantity of ACE and DPP IV inhibitor fragments. All identified sequences exhibit ACE and DPP IV inhibitory activities (Table 1). The ACE inhibitory activity was more pronounced than the DPP IV inhibitory effect despite a lower fragment count (Table 2). The hydrolysate produced by releasing a greater number of peptides with weaker bioactivity may be more potent than the hydrolysate produced by releasing a single peptide with powerful effects [77]. Besides, the EC50 value must be known to calculate discriminant ‘B’. It is entered into the BIOPEP bioactive peptide database once this value has been established. The calculation of ‘B’ is not carried out if the EC50 value is unknown. All of the aforementioned protein evaluation discriminants are listed on the website and were calculated automatically once a protein has been chosen for study [19]. The findings are consistent with in vitro analysis, where four seaweeds (S. binderi, P. sulcata, T. conoides and H. macroloba) were reported to release DPP IV inhibitor [78]. An in silico research revealed that the ribulose-1,5-bisphosphate carboxylase/oxygenase of Caulerpa spp. predominantly consists of antihypertensive peptides [34]. In addition, G. salicornia extract was identified as a potential source of antibacterial peptide in an in vitro and in silico study [79].
Our results indicate that phycoerythrins from the selected rhodophytes mostly contained antihypertensive and antidiabetic effective peptides. Hypertension is a risk factor for cardiovascular diseases [80, 81]. A dipeptidyl carboxypeptidase, which is also known as an angiotensin-converting enzyme (ACE) has a crucial role in protection from hypertension [80]. In the renin-angiotensin system (RAS), angiotensin-I is converted to potential vasoconstrictor angiotensin-II by ACE [82]. Despite being involved in numerous biological processes, serine protease dipeptidyl peptidase-IV (DPP-IV) plays a crucial part in the mechanism of insulin secretion [31]. The blood glucose level and insulin secretion are kept steady by DPP-IV inhibition. Thus, DPP-IV inhibitor peptides would possibly have a therapeutic effect on type 2 diabetes [83]. In addition, one of the important members of Pyropia spp., P. yezoensis is known to afford protective effects against UV and have antioxidant [84-86], anticancer [87], anti-inflammatory [88, 89], antihypertensive [90], and tissue-healing properties [91-93].
Very few studies have reported in-silico analysis on phycoerythrin proteins to produce bioactive peptides. A previous in silico study on phycoerythrin from Gracilaria changii showed the frequency of occurrence (A) of ACE and DPP IV inhibitor was 0.494 and 0.628 [74]. However, the BIOPEP database is increasing, so that value may change. Three groups of values for parameter A were identified: major (≥0.50), moderate (0.100 to 0.499), and small (0.000 to 0.099). For instance, major A assumes that peptides with a specific activity match at least half of a protein chain, which is determined by the ratio of the number of amino acid residues that make up peptides to the length of the protein chain. The major A indicates that there is a high likelihood that the protein will release peptides with this activity by enzymatic processes [20]. This study showed that the frequency of occurrence of ACE and DPP IV inhibitors had much more potential than other activities (Table 2). In contrast, the brown seaweed Fucus vesiculosus extract was shown to be a potent inhibitor of α-glucosidase [94]. In the present study, phycoerythrins derived from rhodophytes were not significant prospective sources of α-glucosidase inhibitors.
When the frequency of occurrence parameter is critical for determining a protein's potential bioactivity, the availability of these confined peptides is linked to peptide bond cleavage. Protein-specific enzymes aid in the production of a large number of peptides with specialized functions. Proteinase K and proteinase P1 were examined for their substrate specifies, and it was discovered that proteinase K can release more bioactive fragments than proteinase P1 [77]. The activity of ACE inhibitors is linked to amino acids with branched side chains. Branched aliphatic amino acids are found at the N-terminus of most ACE inhibitor peptides, with Phe, Trp, Tyr, or Pro at the C-terminus [95]. Proline presented in the C-terminus also showed the DPP IV inhibitory activities [96]. Thus, proline is an important amino acid. High glycine content was evaluated to be potential for the bioactive peptide release. Besides, hydrophobic alanine and glycine presented in the N-terminus showed the potential activity of renin inhibition [97].
Various bioactive peptides were identified in this current study using artificial proteolysis, which has been reported in other scientific literature (Table 5). In-vitro study, the di peptide ‘VF’ was found to be effective as ACE inhibitory activity with the IC50 9.2µM isolated from sardine muscle applied by alkaline protease. The peptide is predicted to be found by papain hydrolysis from phycoerythrin [98]. The peptide ‘DR’ was found effective against ACE activities by molecular docking [40], which can be found in papain, bromelain, and trypsin proteolysis from phycoerythrin. ‘DR’ also possessed the DPP IV inhibitory activity with the IC50 0.00 µM [39]. The peptideRanker is not always a trustworthy source for peptide identification. The bioinformatics tool is unreliable as well, it only means that they still need to advance and expand. For this purpose, it is crucial to keep creating in vitro and in vivo experiments that relate peptide primary and secondary structures to various biological properties [99].
CONCLUSION
In-silico techniques are employed to reduce the lengthy procedure of screening new bioactive peptides from the rhodophytes. These techniques predict the probable bioactive peptides in protein hydrolysates. This in silico study revealed that members of the rhodophytes especially examined could be a valuable source of bioactive peptides for therapeutic benefits. Most of the released bioactive peptides exhibited inhibitory effects on ACE and DPP-IV. Due to their therapeutic benefits against hypertension and type 2 diabetes, members of these protein sequences containing species could be examined in the formulation of functional food or pharmaceutical products. This could introduce a new dimension to the economy by utilizing underutilized resources. The in vitro study will open up a new research way that will enable the exploitation of bioactive peptides in a more beneficial manner. This will be achieved by utilizing appropriate methodologies to extract peptides based on the in silico data.
AUTHORS’ CONTRIBUTIONS
W.L.W: Study conception and design were contributed by; K.N.S: Data analysis and interpretation of results was provided by; K.L: manuscript draft was presented by.
LIST OF ABBREVIATIONS
| NCBI | = National Centre for Biotechnology Information |
| ACE | = Angiotensin Converting Enzyme |
| RAS | = Renin-angiotensin System |
AVAILABILITY OF DATA AND MATERIALS
All the data and supporting information are provided within the article.
ACKNOWLEDGMENTS
Declared none.
APPENDIX A
| Value of a | ||||
|---|---|---|---|---|
| Activity |
Porphyridium purpureum (YP_008965769.1) |
Porphyra purpurea (NP_053978.1) |
Helminthocladia australis (YP_009313668.1) |
Gracilariopsis chorda (YP_009294675.1) |
| AH | 0.5183 | 0.5122 | 0.4463 | 0.4124 |
| APR | 0.0061 | 0.0122 | 0.0169 | 0.0113 |
| GLUI | 0.0366 | 0.0366 | 0.0169 | 0.0113 |
| AI | 0.0061 | 0.0061 | - | - |
| AM | 0.0122 | 0.0122 | 0.0113 | 0.0113 |
| AB | - | - | 0.0056 | 0.0056 |
| AC | - | - | 0.0113 | 0.0056 |
| AO | 0.0671 | 0.0488 | 0.0452 | 0.0395 |
| AT | 0.0122 | 0.0122 | 0.0113 | 0.0113 |
| 35PD | 0.0122 | 0.0061 | 0.0056 | |
| AVI | - | - | 0.0056 | 0.0056 |
| LIG | - | - | 0.0056 | - |
| DPP3 | 0.0915 | 0.0915 | 0.0847 | 0.0847 |
| DPP | 0.622 | 0.628 | 0.6271 | 0.5763 |
| HYPL | 0.0061 | - | - | - |
| HE | - | - | 0.0056 | 0.0056 |
| HYP | 0.0244 | 0.0305 | 0.0339 | 0.0226 |
| LCP | 0.0061 | - | - | - |
| LEUT | - | - | 0.0056 | 0.0056 |
| NE | 0.0305 | 0.0183 | 0.0113 | 0.0113 |
| PAM | 0.0061 | 0.0061 | 0.0113 | 0.0113 |
| RE | 0.0183 | 0.0244 | 0.0113 | 0.0113 |
| REN | 0.0122 | 0.0122 | 0.0339 | 0.0282 |
| ST | 0.0183 | 0.0244 | 0.0452 | 0.0452 |
| TOX | - | - | 0.0056 | - |
| XAAP | 0.0061 | - | - | - |


